※ GPS-SNO INTRODUCTION:
The 1998 Nobel Prize in Physiology or Medicine was awarded for seminal discoveries that nitric oxide (NO) is a freely-diffusible signalling molecule and second messenger, regulates the production of cyclic GMP (cGMP), and plays essential roles in the cardiovascular system. Later, flood of studies challenged this fundamental view by observing that NO could spatially and temporally target specific cysteine thiols and transition metals of proteins, a reversibly post-translational modification (PTM) termed as S-nitrosylation (Foster, et al., 2009; Foster, et al., 2003; Hess, et al., 2005; Hess, et al., 2001; Stamler, et al., 2001; Tannenbaum and White, 2006). Although enzymatic mechanisms of protein S-nitrosylation were still elusive, several enzymes were proven to facilitate S-nitrosylation or de-nitrosylation reactions. For example, Cu, Zn superoxide dismutase (SOD) and thioredoxin (TRX) could promote S-nitrosylation, while protein disulfide isomerase (PDI) might regulate de-nitrosylation (Hess, et al., 2005; Tannenbaum and White, 2006). Current progresses proposed that S-nitrosylation could modulate proteins' stabilities (Li, et al., 2007), activities (Tsang, et al., 2009) and trafficking (Hernlund, et al., 2009; Ozawa, et al., 2008), and play important roles in a variety of biological processes, including transcriptional regulation (Li, et al., 2007), cell signalling (Whalen, et al., 2007), apoptosis (Tsang, et al., 2009), chromatin remodeling (Nott, et al., 2008) and so on. Moreover, aberrant S-nitrosylation has been implicated in numerous diseases and cancers (Foster, et al., 2009; Foster, et al., 2003; Tsang, et al., 2009). In this regard, experimental identification of S-nitrosylated proteins with their sites will be a foundation of understanding the molecular mechanisms and regulatory roles of S-nitrosylation.
In this work, we manually collected 467 experimentally verified S-nitrosylation sites in 302 unique proteins from scientific literature. Previously, we developed an algorithm of GPS 2.0 (Group-based Prediction System) for prediction of kinase-specific phosphorylation sites (Xue, et al., 2008). Here, we greatly improved the method and released GPS 3.0 algorithm. Then we developed a novel computational software of GPS-SNO 1.0 for prediction of S-nitrosylation sites. The leave-one-out validation and 4-, 6-, 8-, 10-fold cross-validations were calculated to evaluate the prediction performance and system robustness. By comparison, the performance of GPS 3.0 algorithm was much better than several other approaches, with an accuracy of 75.70%, a sensitivity of 55.32% and a specificity of 80.11% under the low threshold. As applications of GPS-SNO 1.0, we also collected 485 potentially S-nitrosylated substrates from PubMed. These proteins were detected from large-scale or small-scale studies, while the exact S-nitrosylation sites were still not experimentally determined. Successfully, we predicted 371 (~76%) of these targets with at lease one potential S-nitrosylation site. These prediction results might be a useful reservoir for further experimental verification. Finally, the online service and local packages of GPS-SNO 1.0 were implemented in JAVA 1.4.2 and freely available at: http://sno.biocuckoo.org/.
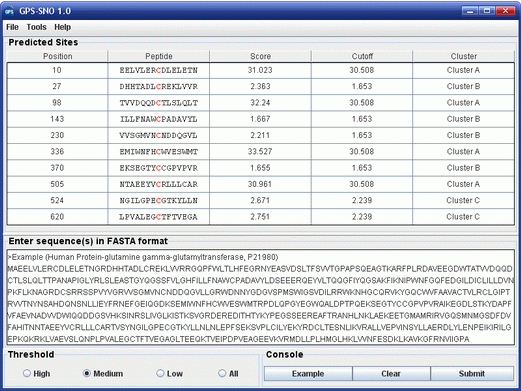
GPS-SNO 1.0 User Interface
For publication of results please cite the following article:  GPS-SNO: Computational prediction of protein S-nitrosylation
sites with a modified GPS algorithm. Yu
Xue, Zexian Liu,
Xinjiao Gao, Changjiang Jin, Longping Wen, Xuebiao Yao, and Jian Ren. Plos One. 2010;5(6):e11290
GPS-SNO: Computational prediction of protein S-nitrosylation
sites with a modified GPS algorithm. Yu
Xue, Zexian Liu,
Xinjiao Gao, Changjiang Jin, Longping Wen, Xuebiao Yao, and Jian Ren. Plos One. 2010;5(6):e11290
|



